Global Site
Breadcrumb navigation
Graph-based Relational Learning in Healthcare
Featured TechnologiesJanuary 30, 2020

Introduction
The world we live in is a graph – everything is connected.
We see the world as a collection of entities (e.g., people, cities, devices, molecules) and their relations to each other. In addition to relations, each entity has attributes which characterize it; such characterizing attributes are in nature multi-modal, i.e., each entity can be associated with various attributes coming from multiple data modalities (e.g., numerical, text, visual).
Examples are:
- in social activities, people are connected to each other by multiple relations (e.g., work relations, family relations, social relations); each person has numerical attributes (e.g., age, height), text attributes (e.g., their work curriculum, their medical diagnosis, their social posts) and visual ones (e.g., a picture of their face);
- in chemistry, molecules can be connected to each other by multiple relationships (e.g., protein known interactions, common protein targets)
- different entity types can also be connected to each other with relations
- e.g., people and devices are connected to each other: a mobile phone, a car, a house belonging to a certain person (each device has in turn numerical, text and visual attributes)
- cities belong to a certain country (each city has in turn numerical, e.g., latitudes and longitudes, number of crimes in the city; textual, description of the city; and visual, pictures of places in the city)
By analyzing such once thought to be fragmented data, now we can reveal hidden relationships and create new business values.
Goals and Challenges
Extracting actionable insights from these multi-relational, multi-modal knowledge graphs is our ultimate goal.
We want to reach a final state where we can provide our customers a valuable intelligent system that enables to give them answers to very specific questions like “What is the best set of therapies for this patient”?
At the same time we want to enrich our answers with a human interpretable explanation like “The best set of therapies for this patient are X and Y because their combination has already resulted in positive outcomes and no adverse events in persons with similar characteristics”
The machine learning challenges to reach these goals are numerous:
- extracting and integrating multi-modal information from unstructured and structured data;
- working with missing, biased and protected knowledge (missing labels, missing features, and biased and protected data);
- bringing together the multiple modalities and the multiple relationships into a suitable representation that allows us to learn a machine learning model embedding the full potential of the knowledge we have available;
- scaling models to very large numbers of entities, attributes and relations and can answer in real time if required.
Our technical approach
Towards the achievement of our goals, we tackle the challenges mentioned above with this technical approach:
- Integrate and induce a relational and multi-modal representation graph by integrating structured and unstructured knowledge into a multi-modal multi-relational graph;
- Learn complete neural-relational models for the representations that allow to perform downstream machine learning task like classification, prediction and regression
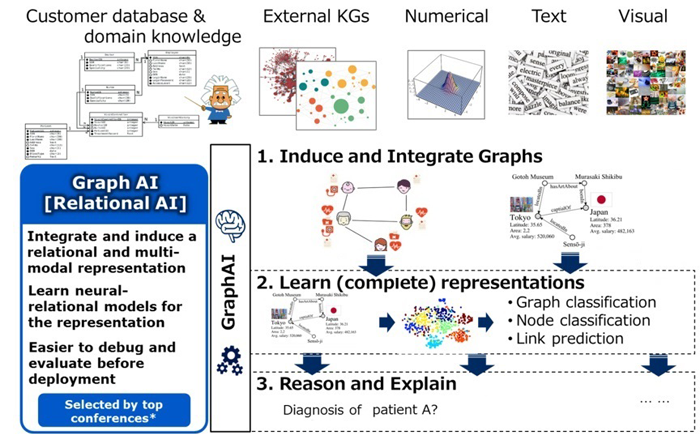
Graph-based Relational Learning in HealthTech
Use case in precision medicine
With new biomarkers, physicians can provide better care for cancer patients. They can increase the survival rates and avoid unnecessary complications during treatments.
In immuno-oncology, treating physicians are aiming at:
- designing the best treatment for cancer patients with respect to the survival and disease progress;
- predicting severe side-effects that would endanger patient’s life and require to stop with the treatment;
Our Graph-based Relational Learning provides positive patient outcomes which are the ultimate measure of doctors’ success and bring unmeasurable value to the society:
- increased survival rates due to better treatment selection;
- fewer health complications during treatments.
NEC is collaborating with several academic and industry partners on several projects which demonstrate the value of Graph-based Relational Learning in helping physicians achieve their aims in immune-oncology.
In one project, the goal is to find the target site for the design of vaccines, which induce an immune response to suppress cancer in patients. We first use bioinformatic approaches to identify candidate sites based on DNA sequencing in the patient. We then proceed with our graph-based relational learning approach.
For this problem, we construct a graph in which each node corresponds to a target site candidate; we connect nodes with edges based on their similarity. Further, we annotate each node with known biological measured data and other known biological background knowledge; however, all biological measured data are not available for all target sites. Thus, we have missing data. We can build the strong representations of the target site by using our graph-based relational learning even though there are missing data. These representations of the target site serve as inputs for downstream approaches to determine the quality of each target site for the individual patient.
